Blogs & News
Aridhia & ICODA Workbench – Grand Challenges, Driver Projects and Beyond
The COVID-19 pandemic brought both health and inequalities into sharp focus. From the early stages of the pandemic, it was clear that some demographics were more affected than others.
The primary mission for the International COVID-19 Data Research Alliance (ICODA) was to build an open and trustworthy international research partnership to support a rapid response to COVID-19 and a long-term alliance for making data accessible to health researchers and scientists around the world. The ICODA Workbench strived to unite international health research data to enable discoveries that benefit everyone, everywhere, by reducing the harm of COVID-19; and enabling an efficient data response to future pandemics and other health challenges.
The ICODA Driver projects and ICODA Grand Challenge Initiative awards cast a wide net both in terms of geography and the nature of the questions being asked. The variety of research questions, all of which related to the COVID-19 pandemic, meant that each Driver Project and Grand Challenge project had its own characteristics and specific requirements. In the spirit of open science, all the outputs and tools that we co-created were made publicly available by the project teams. Metadata was made findable and accessible via FAIR Data Services and syndicated to the ICODA Health Data Research UK (HDR UK) Gateway.
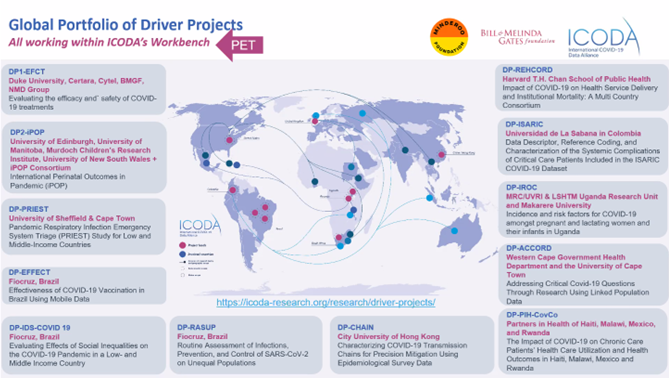
Aridhia has spent over 18 months providing their Digital Research Environment (DRE) (a more complex system of what is usually referred to as a Trusted Research Environment (TRE)) for ICODA Driver Projects, COVID-19 Grand Challenge projects, and another DRE environment for the International Severe Acute Respiratory and emerging Infection Consortium (ISARIC) at Oxford University. During this time, we have maintained two Workbench Hubs (one in Western Europe and one in North Europe) including 15 workspaces and FAIR datasets for ICODA Driver Projects 1 and 2, and 16 workspaces and FAIR datasets for the ICODA Grand Challenge.
Through the ICODA programme the ICODA Workbench demonstrated that COVID-19 was a truly global pandemic and that wherever healthcare is happening, health research should also be happening as a matter of routine. Utilising a cloud-based workbench, ICODA championed how we can’t let useful data and insights go to waste and we can’t allow the location of that data to affect unconscious bias in those studies, particularly when we’re dealing with a new disease.
You can see the full list of publications here.
For all the diversity of the ICODA projects, there was a common denominator and that is the approach to how data, metadata, analysis, and insights are produced to ensure widespread access, re-use, and sharing of data and methods. All the projects made use of a subscription to the ICODA Workbench to enable a research lifecycle that’s consistent with the principles of open science and the safeguarding of privileged data and content.
Driver Projects
We previously reported on several of the projects involved in the Grand Challenge event during its run.
“My research team used the FAIR database and Workbench to share data and computer codes, respectively. The platform provided a very good place for my remote-working team to share information and standardize their working processes. We appreciate the help from your team in the past year!”
Dr Xiaofan Liu, City University of Hong Kong
We followed up with Juliane Oliveira from Fiocruz and her team who were seeking to find ways to mitigate SARS-CoV-2 spread, and saw in their final report some of the tangible results of their hard work. Among other things, they established the Brazilian COVID-19 data lake from various sources relating to infections, vaccinations, government interventions, human mobility, and population inequality. Community-level aggregated datasets were created off the back of this, allowing for real-time assessment of the pandemic in over five and a half thousand municipalities, fostering community involvement. The team also create the Platform for Analytical Models in Epidemiology (PAMEpi). This platform aims to provide a user-friendly interface, enabling researchers and policymakers to assess the impact of the spread of (emerging) infectious diseases in their respective areas of interest.
Dr Xiaofan Liu, an expert in AI and mathematical modelling, led a team looking at COVID-19 transmission chains. He spoke with us about his team’s use of the DRE. You can find more information and papers relating to that work here.
The ICODA Workbench
The ICODA Workbench was built from the Aridhia DRE and follows our approach of creating a research lifecycle that breaks down many of the barriers to enabling open science programs. We lean heavily on our partners at Microsoft for running the DRE on Azure and are extremely grateful to the Microsoft AI for Good program for granting Azure credits for the Grand Challenge projects. Azure allows us to build effective collaboration networks across the world, linking many data contributors to clinical, scientific, and data science users.
Our primary goal is to enable research teams and data controllers to focus on their science and not on the underlying data management, collaboration and governance infrastructure. The DRE allows data controllers to publish their metadata and other useful artefacts openly, gives them control on the management of data access requests, and provides visibility of how their data is being used. Clinical, scientific and data science users have the freedom to operate on requested data in safe, secure analytical Workspaces, and mint DOI’s that give visibility to methods and code, allowing other teams to build on top of previous work rather than starting from scratch.
November 1, 2022
Eoghan Forde
Eoghan Forde is a Project Manager with Aridhia. He has a background in Biomedical and Clinical Sciences, having completed his PhD in Precision Medicine at the University of Edinburgh Medical School. Since joining Aridhia in October 2021, he has been providing subject matter support for Genomics and Bioinformatics within the DRE.