Blogs & News
Working with the OMOP CDM in the Aridhia DRE
The utilisation of observational healthcare data has become increasingly important in evidence-based medicine and healthcare research. Researchers, clinicians, and policymakers leverage this data to study disease patterns, treatment effectiveness, patient outcomes, and healthcare utilisation on a large scale. However, the heterogeneity and variability in data sources, formats, and standards pose significant challenges to the meaningful analysis and interpretation of observational healthcare data.
This is where a Common Data Model (CDM) becomes crucial. A CDM is a standardised framework that structures and consistently organises observational healthcare data, facilitating interoperability and comparability across different datasets.
What is the OMOP CDM?
One of these CDMs is The Observational Medical Outcomes Partnership Common Data Model, commonly known as the OMOP CDM, which is designed to organise and harmonise healthcare data for observational research. Developed by the Observational Health Data Sciences and Informatics (OHDSI) collaborative, the OMOP CDM provides a common language and structure for representing diverse healthcare data sources, enabling researchers to conduct large-scale analyses across different datasets.
At its core, the OMOP CDM defines a set of tables and relationships that standardise the representation of clinical data, such as patient demographics, medical conditions, treatments, and outcomes. This standardised format allows researchers to seamlessly integrate data from various healthcare databases, regardless of their source or format. By ensuring a consistent structure, the OMOP CDM promotes interoperability and facilitates the pooling of data for robust observational studies and analyses.
The Challenges of working with the OMOP CDM
Complexity and Variability
Healthcare databases often differ in terms of data structure, coding systems, and clinical practices, making it challenging to harmonise data across diverse sources. The process of mapping and standardising these varied data elements to the OMOP CDM requires meticulous attention to detail and a deep understanding of clinical concepts.
Interpreting Results
Observational studies inherently introduce biases, confounding variables, and other complexities that differ from the controlled environment of clinical trials. Researchers working with OMOP data must carefully consider these nuances to draw accurate conclusions and make informed decisions, especially when the stakes involve public health or regulatory considerations.
Volume and Granularity
OMOP databases encompass a vast array of patient information, including electronic health records, claims data, and other real-world evidence. Managing and analysing such extensive datasets demands robust computational infrastructure and sophisticated analytical techniques. Researchers must grapple with issues related to data storage, processing speed, and the scalability of their analytical methods to extract meaningful insights from these rich datasets efficiently.
Overcoming these Challenges with the Aridhia DRE
The Aridhia DRE provides a secure and scalable platform for data ingestion, transformation, validation, and quality control, making it the ideal platform to support the entire OMOP journey all the way through to data analysis. Aridhia are a European Health Data and Evidence Network (EHDEN) SME, and supports the OMOP ETL journey for a number of DRE customers, including Great Ormond Street Hospital and the Sydney Children’s Hospital Network.
To transform health records into an OMOP source, a series of ETL (extract, transform, load) pipelines are needed. These pipelines involve:
- • Extracting the raw data from the source systems, such as electronic health records, claims databases, registries, or surveys.
- • Transforming the data into the OMOP common data model (CDM), which defines a set of standard tables and fields for storing health data.
- • Loading the data into a relational database or a cloud storage service that supports SQL queries.
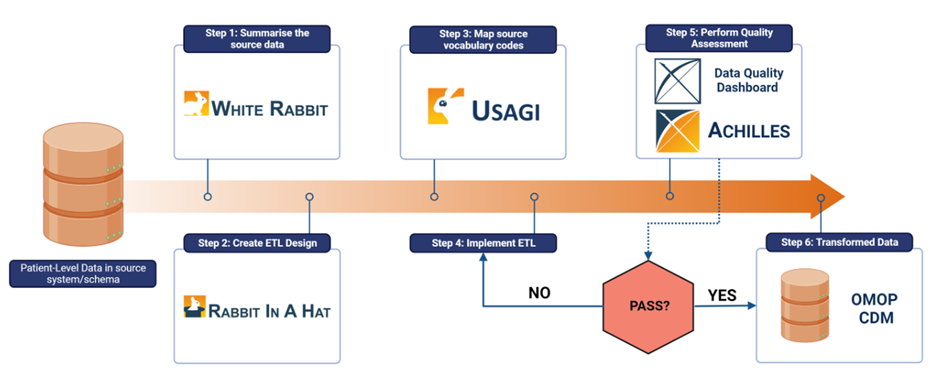
The step-by-step process for mapping data sources to the Observational Medical Outcomes Partnership Common Data Model (OMOP CDM), using OHDSI software tools. ETL: Extraction, Transformation and Load; DQD: Data Quality Dashboard. Reproduced using [1]
The technologies that can be used to implement these ETL pipelines vary depending on the source and target systems, but some common ones are:
- • Python or R scripts for data manipulation and transformation
- • SQL scripts for data validation and quality checks
- • Apache Airflow or Luigi for workflow orchestration and scheduling
- • PostgreSQL or Microsoft SQL Server for relational database management
The Aridhia DRE allows data owners to share their OMOP data with authorised researchers, who can access and analyse the data using a variety of tools and methods within the DRE:
- • Metadata management: OMOP datasets are fully supported in our native metadata catalogue FAIR Data Services.
- • Cohort definition and characterisation: Researchers can define and compare groups of patients based on their clinical features, exposures, outcomes, or other criteria using the OHDSI Atlas tool.
- • Population-level effect estimation: Researchers can estimate the causal effects of interventions or exposures on outcomes using the OHDSI CohortMethod package.
- • Patient-level prediction: Researchers can build and validate predictive models for individual patient outcomes using the OHDSI PatientLevelPrediction package.
Each Aridhia DRE workspace provides an out-of-the box R Development Environment and Jupyter Notebook application, without the need to use a virtual machine, saving on platform costs while providing researchers with the tools they are familiar with, along with in-built data analysis modules and a no-code SQL development environment. OMOP data can be large and a zero-transfer approach to working with this data can be enabled, allowing for direct read-only access to approved cohorts of OMOP data from workspace applications, including specialist R web applications.
While OMOP and its Common Data Model offer a promising avenue for advancing observational research in healthcare, researchers must navigate a landscape fraught with challenges. Addressing issues related to data harmonisation, scalability, and result interpretation is crucial for unlocking the full potential of OMOP data and realising its impact on improving patient outcomes and healthcare delivery. The Aridhia DRE provides a secure, scalable end-to-end platform to support the entire OMOP data journey, from transformation, to analysis.
February 12, 2024
Eoghan Forde
Eoghan Forde is a Project Manager with Aridhia. He has a background in Biomedical and Clinical Sciences, having completed his PhD in Precision Medicine at the University of Edinburgh Medical School. Since joining Aridhia in October 2021, he has been providing subject matter support for Genomics and Bioinformatics within the DRE.
